Running AmberTools 1.4 and Amber 11 on Microsoft Windows
| General remarks |
Installation on Windows using Cygwin |
|
The performance of the native Windows executable of PMEMD has
been tested on the Windows HPC benchmarking cluster Rhiannon
located at the
Microsoft Enterprise Engineering Center.
All benchmarks have been performed by
Thorsten Wölfle,
Andreas W. Götz and
Ross C. Walker
(WMD lab, SDSC).
Machine Specs (Rhiannon)64 IBM iDataPlex DX360 M2 compute nodesIntel Nehalem Xeon 5500 series (total 512 cores) 24GB DDR3 RAM per node Six 36-port QDR InfiniBand switches, 72 QDR InfiniBand Adapters Copper and optical cables for a fully connected non-blocking QDR IB fabric
Windows HPC Server 2008 V2 SP1 AMBER 11 PMEMD compiled with Intel Cluster Toolkit 3.2 List of benchmarksYou can download the Amber11 Benchmark Suite used in this work here. This suite covers the range of sizes and parameters used in common, explicit-solvent MD simulations with PMEMD. Results for three benchmarks are reported below.1) JAC NVT = 23,558 atoms 2) Factor IX NVT = 90,906 atoms 3) Cellulose NVT = 408,609 atoms You can download a Python script which can be used as a template to run the test suite on a Microsoft HPC cluster. Download it from here, copy it to the Amber11 Benchmark Suite directory and edit it as appropriate. Use this as follows:
CYGWIN> cp test_pmemd_win.py Amber11_Benchmark_Suite/PME This will generate batch scripts for the submission of the benchmarks on up to 32 nodes using 8 cores per node and up to 64 nodes using 4 cores per node. Submit the jobs by executing the batch script now:
DOS> cd Amber11_Benchmark_Suite |
|
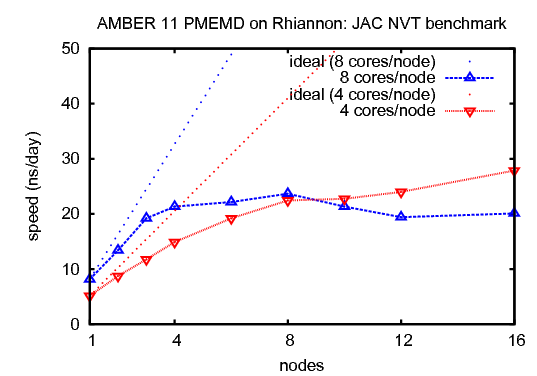
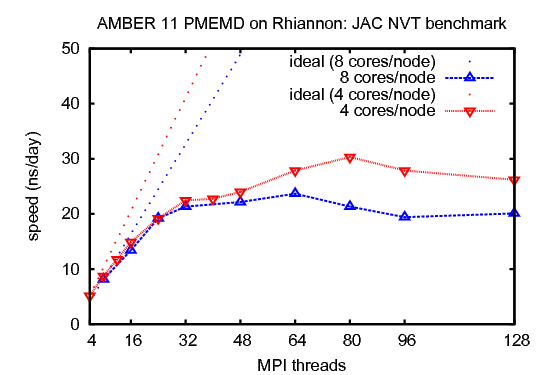
1) JAC (Joint Amber CHARMM) production benchmark for dihydrofolate reductase (DHFR), NVTThis benchmark consists of 23,558 atoms including water. It has been updated from the original JAC benchmark to include more realistic simulation parameters that reflect production use.&cntrl |
|
|
|
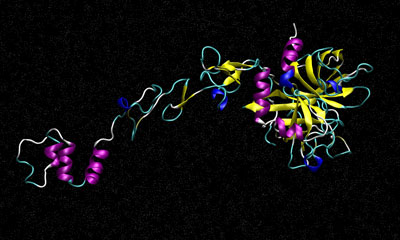
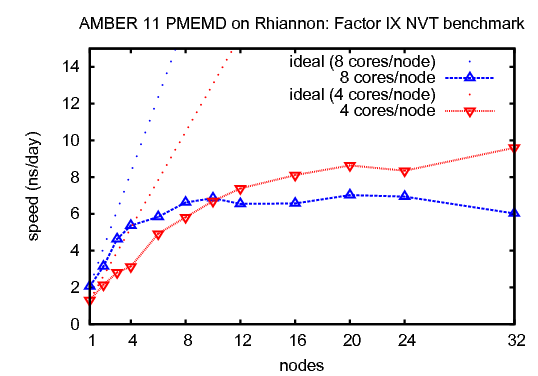
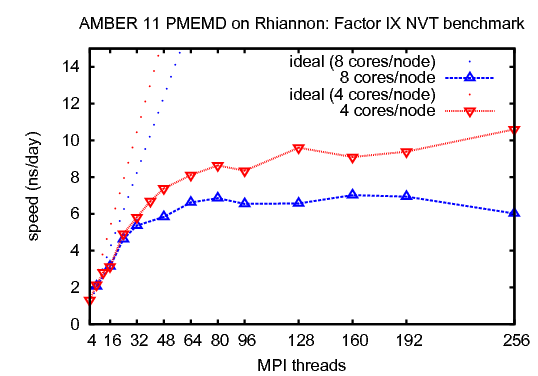
2) Factor IX, NVTThis benchmark simulates the blood coagulation factor IX. It consists of 90,906 atoms including water.&cntrl |
|
|
|
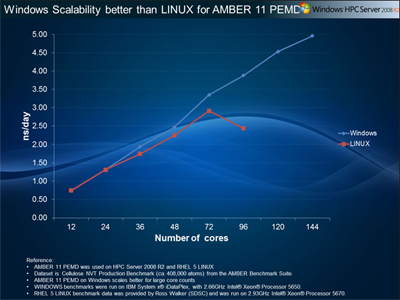
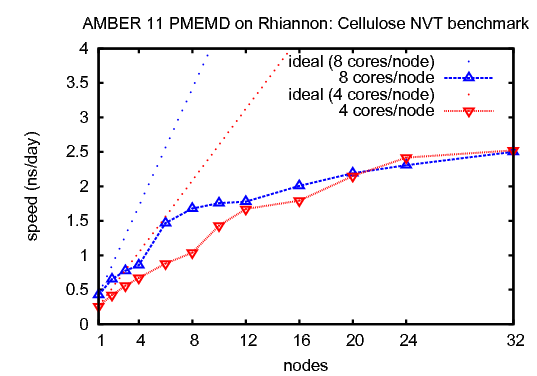
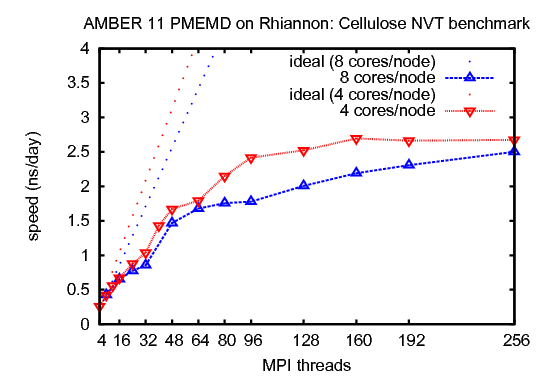
3) Cellulose, NVTThis benchmark simulates a cellulose fiber. It consists of 408,609 atoms including water.&cntrl |

|
|
|
| last modification: 2010/06/01 |